Regulation of a nutrient-dependent cell growth
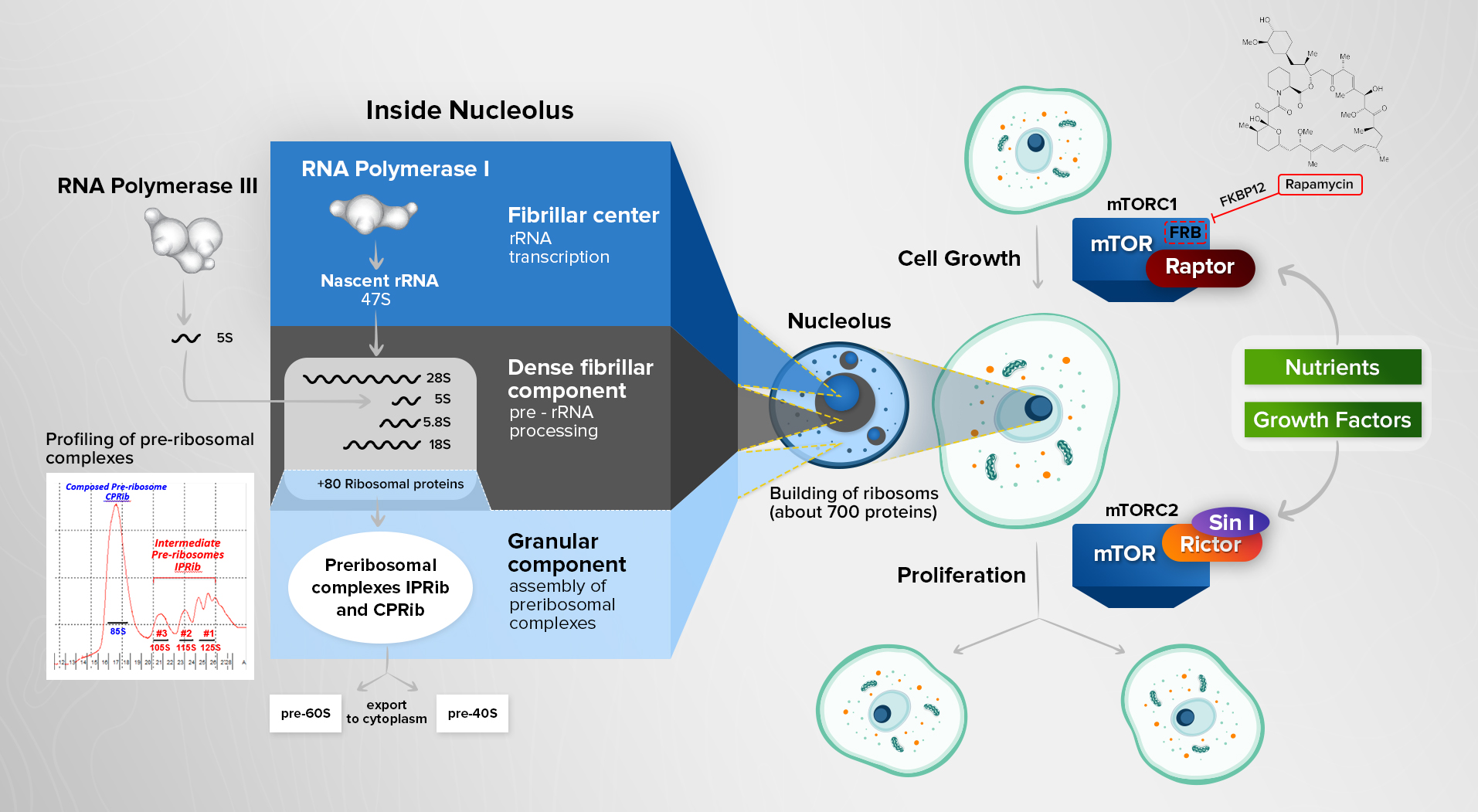
Origin of eukaryotes is a fundamental breakthrough in evolution. By enhancing aerobic gain of energy, eukaryotic cells transformed a complexity of biological systems marked by a substantial increase in cell size and emergence of multicellular organisms. If quantified, an average mammalian cell is at least one thousand times larger than a bacterial E. coli. A substantial increase of eukaryotic cell size obligates a coordination of cell growth with metabolic input. This coordination is installed in every eukaryotic cell by a stringent signaling pathway controlling accumulation of cellular mass according to nutrients available to cell. It is known as a target of rapamycin (TOR) pathway and is referred as mTOR in mammalian cells. A central component of this pathway is a TOR protein that is an essential and large serine/threonine kinase conserved in all eukaryotes. It has been named after a natural compound rapamycin known as a highly specific allosteric inhibitor of TOR that was selected originally for its growth suppressing activity in yeast. Rapamycin is secreted by bacteria Streptomycin hygroscopicus and is likely an advantageous trait in territorial competition with yeast.
The biochemical studies determined that mTOR exists in two distinct complexes distinguished not only functionally but also by their sensitivity to rapamycin. mTOR in a complex with raptor forms the mTOR Complex 1 (mTORC1). It has been defined as a nutrient-dependent complex controlling cell growth (accumulation of cellular mass) by regulating a rate of protein synthesis through phosphorylation of its substrates 4EBP1 and the kinases of S6 ribosomal protein S6K1 and S6K2 (the members of AGC kinase family). Nutrients and growth factors promote protein synthesis by regulating a kinase activity of mTORC1. Interestingly, lysosomes were identified as the amino acid sensing organelle. Amino acids control a basal activity of mTORC1 by its translocation on a lysosomal surface mediated by Rag GTPase proteins. Growth factors enhance a basal kinase activity of mTORC1 by regulating Rheb GTPase. Deprivation of amino acids inhibits activity of mTORC1 leading to decline of protein synthesis, activation of autophagy (an intracellular “self-eating” survival process induced by starvation) and decrease of cell size. A lipophilic macrolide rapamycin mimics a nutrient deprivation effect by targeting mTORC1. Rapamycin in a complex with its small protein receptor FKBP12 inhibits mTORC1 by binding with a high specificity to the FKBP12 and rapamycin binding (FRB) domain of mTOR. FRB represents a stretch of 100 amino acids adjacent to the kinase domain which is located at the C-terminus of mTOR. A single mutation within FRB domain prevents binding of the rapamycin/FRB drug complex on mTOR and renders cells insensitive to rapamycin.
A binding of rictor and SIN1 proteins to mTOR forms a distinct complex without raptor known as mTORC2. The kinase activity of mTORC2 depends on a growth factor signaling and it regulates cell proliferation, metabolism and migration by functioning as a regulatory kinase of other AGC kinases (Akt, PKC and SGK) by phosphorylating their hydrophobic motifs. A basal activity of mTORC2 is growth factor independent and maintains a constitutive phosphorylation of the turn motifs of its kinase substrates. A basal activity of mTORC2 is sensitive to ATP depletion and observed under glucose deprivation. In a contrary to mTORC1, mTORC2 is not targeted by rapamycin suggesting that the FRB domain is hidden by rictor and SIN1 bound to mTOR. Besides, mTORC2 is less conserved compare to mTORC1 because it exists in yeast and animals but it has not been found in plants. Thus, two distinct mTOR complexes defining the fundamental roles of mTOR signaling in regulation of cell growth and proliferation indicate that mTOR determines growth of organism in development and maintains its body size according to a nutrient consumption and environmental factors. Deregulation of mTOR signaling is common in human diseases.
Current lab interests:
1. Regulation of ribosomal assembly in mammalian cells and its deregulation in cancer.
Ribosomal biogenesis is a cellular biosynthetic process of constructing the massive ribonucleoprotein (RNP) complexes known as ribosomes. A rate of ribosomal assembly determines a cellular protein synthesis and cell growth. In eukaryotes, a designated sub-nuclear organelle nucleolus has been evolved to accommodate an intensive ribosomal biogenesis. Nucleolus is a non-membrane bound organelle located in the nucleus with a defined proteome of about 700 proteins including various ribosomal biogenesis factors. It is visualized as a dense particle at the chromatin sites of multiple tandem ribosomal DNA (rDNA) repeats. Building of ribosomes is a main functional role of nucleolus. It contains distinct functional sections (fibrillar center, dense fibrillar component and granular component) to accommodate the complex process of building of ribosomes. The rDNA repeats are actively transcribed by RNA polymerase I into a nascent rRNA of 13 kb in size at the fibrillar center. A processing of rRNAs takes place at the dense fibrillar component. Pre-ribosomal complexes get assembled at granular component.
Ribosomal biogenesis regulates cell growth and it is controlled by a nutrient dependent mTOR signaling. To study regulation of ribosomal assembly, we isolated and characterized native pre-ribosomal complexes and identified the Intermediate Preribosomes (IPRibs) and Composed Preribosome (CPRib). It is an original study because most of pre-ribosomal complexes were isolated by performing the recombinant expression and isolation of ribosomal biogenesis factors indicating that native pre-ribosomes were not characterized. IPRibs contain the rRNA modification (methylation, acetylation and pseudouridylation) factors known to play crucial roles in the ribosomal assembly and functional performance of ribosomes. CPRib is an assembled preribosome at its composed state prior to its disintegration to pre-60S and pre-40S subunits with a following export from nucleus to cytoplasm. Regulation of ribosomal assembly is our main research interest with the focus on the following questions:
1. How mTOR signaling regulates the transcriptional activity of RNA polymerase I?
2. How assembly of ribosomes and rRNA modifications are regulated? The attractive question to address is whether the changes in modifications of rRNAs lead to formation of different types of ribosomes that might play a critical role in adaptations to different physiological or environmental conditions.
3. The mechanisms of intensive ribosomal biogenesis in cancer cells leading to an accelerated assembly of ribosomes and coupled with enlargement of nucleoli are yet to be unraveled.
2. Cancer drug development by inducing a cytotoxic oxidative stress.
Nutrient deprivation studies show that cancer cells are highly adaptive to amino acid deprivation by inhibition of mTOR signaling and induction of autophagy. In a contrary, a glucose deprivation also induces autophagy but much more stressful to cancer cells. Screening of different cancer cell types determined that KRAS mutant cells are highly sensitive to glucose deprivation. A glucose starvation of KRAS mutant cancer cells induced a robust generation of reactive oxygen species (ROS) leading to a potent cytotoxic oxidative stress. Mitochondria have been identified as a source of ROS production triggered by a cellular ATP depletion. While working on unraveling of a suicidal ROS generation, we developed a potent oxidizing drug combination selectively killing KRAS mutant cancer cells and its clinical relevance is yet to be determined.
Our research interests in understanding of a cytotoxic oxidative stress in cancer:
1. To determine the mechanisms of a mitochondrial suicidal ROS generation in cancer cells.
2. How ROS induce a pre-apoptotic nuclear condensation and initiate cancer cell death.
3. Cancer drug development by triggering a cytotoxic oxidative stress in cancer cells.




